2011-14
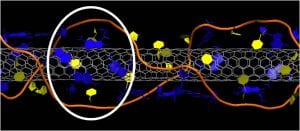
Using extensive molecular dynamics calculations, we studied the structure of DNA on carbon nanotubes. We showed that DNA adopts novel secondary structures including a self-stitching motif that seems effectively to ligate multiple strands into one helix, beta-barrels, and loop-like structures.
See, for example (simulation work led by Jeetain Mittal):
“Molecular Basis of Single-Walled Carbon Nanotube Recognition by Single-Stranded DNA,” Daniel Roxbury, Jeetain Mittal, Anand Jagota(Nanoletters, 12 [3] 1464-1469 (2012))
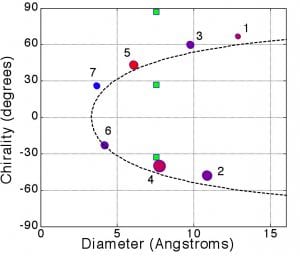
Recently, we have looked at what aspects of DNA/SWCNT interactions allow enantiomer recognition.
Older Work (July 2010)
Molecular simulation of beta-barrel DNA structures on SWNTs support the idea that such secondary structures are stabilized by their interaction with the nanotube core. (Journal of Physical Chemistry C, 2010, Roxbury et al.)
(July 2009)
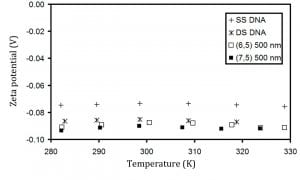
We have determined the charge density of dna-cnt hybrids by capillary electrophoresis measurements and have shown these are consistent with a dna beta-barrel model
(August 2007)
Lateral motion of bases is relatively facile with a 2kT barrier between energy minima.